Alignment of V and J segements with SeaView.
Page in construction…
SeaView is a “multiplatform, graphical user interface for multiple sequence alignment and molecular phylogeny”. In addition to the traditional display of nucleotides, it can color the alignment by codons, or display the DNA sequence translated as protein. This makes it very useful for manual alignemnt of V and J segments.
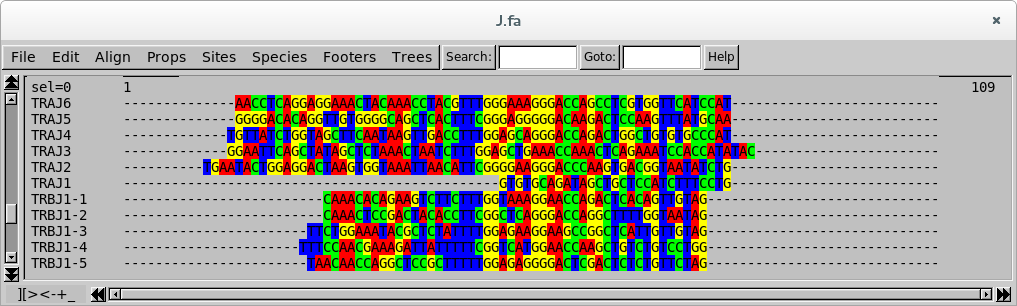
Nucleotide view
J segments must be aligned on the FGxG motif, but in the default view it is
not very obvious to see. Switch to the codon view or the aminoacid view.
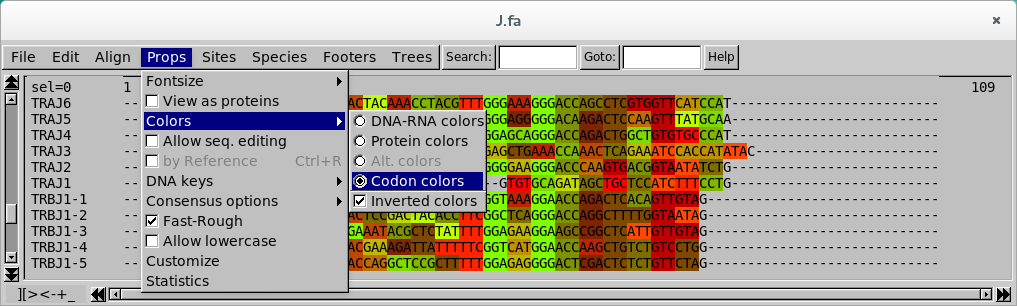
Codon view
Here, the FGxG motif stands out like for example “TTCGGTCATGGA”. This mode is very useful for finding the relevant reading frame, by adding or removing gaps before the first nucleotide.
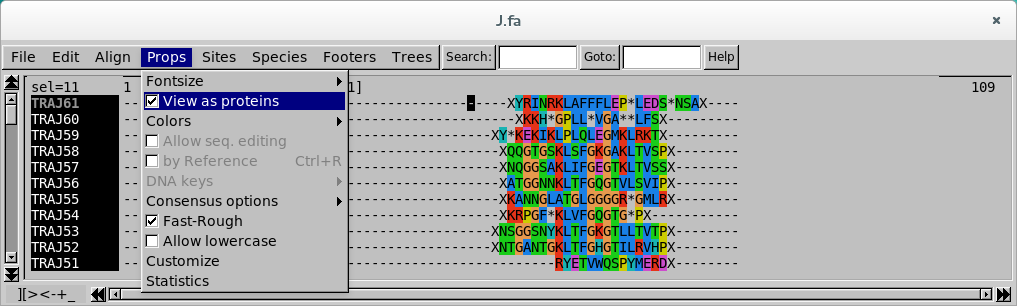
Aminoacid view
This mode is very useful for aligning in-frame sequences, checking the final alignment, and spotting easily the stop codons in pseudogenised segments.